|



| |
Projects

|
|
|
NSF Award
Number |
0727870 |
|
NSF
organization |
CMMI |
|
Program Manager |
Demitris A. Kouris (current
PO of Nano-
and Bio-Mechanics division), past Jimmy Hsia. |
|
Start date |
August 1, 2007 |
|
Expires |
July 31, 2010
(estimated) |
| |
|
|
Investigators |
 |
William A. Goddard,
III (PI) |
 |
Andres
Jaramillo-Botero (Co-PI, Coord.) |
 |
Mario Blanco
(Co-PI) |
 |
Youyong Li
(Co-PI) |
 |
Seung Soon Jang
(Co-PI @ Gatech) |
|
| Abstract |
|
 |
|
 This research will
develop a strategy for using first principles theory and computation to
determine the atomistic details of polymer hydrogel double network
structures applicable in the development of scaffold-supported cell
therapies to promote cartilage regeneration. Recent advances in
first-principles-based molecular simulations that allow the description of
systems with 1,000s-millions of atoms with chemical and structural detail at
the Materials and Processing Simulation Center in the California Institute
of Technology will enable the essential framework to: This research will
develop a strategy for using first principles theory and computation to
determine the atomistic details of polymer hydrogel double network
structures applicable in the development of scaffold-supported cell
therapies to promote cartilage regeneration. Recent advances in
first-principles-based molecular simulations that allow the description of
systems with 1,000s-millions of atoms with chemical and structural detail at
the Materials and Processing Simulation Center in the California Institute
of Technology will enable the essential framework to:
-
simulate the
critical nano bio-mechanical properties of gel polymer networks, including
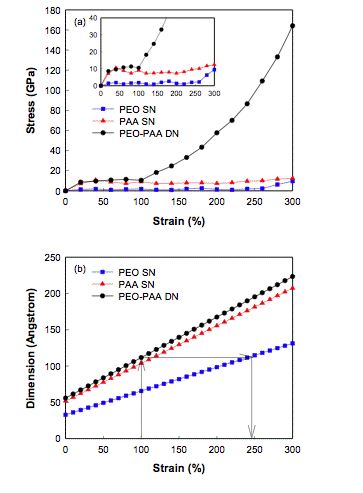
mechanoregulation (figure to the left shows the stress-strain relationship
for a PEO/PAA DN [Seung Soon et al, 2007], and
-
develop an
increased understanding of fundamental mechanisms that regulate in-vivo
performance for the development of new/enhanced materials.
This work will
validate the strategy on prototypical systems and set the stage for
important applications in Tissue Engineering.
This research is critical to improve our understanding of, and to enhance
our ability to emulate the, nano-mechanical properties of natural cartilage.
Cartilage has a limited self-repair capacity and traditional therapies for
musculoskeletal conditions involving cartilaginous tissue have relied on
surgical procedures for full joint replacements when local
repair/replacement is not possible; these methods have proven to be
ineffective in the long-term. Musculoskeletal conditions remain as one of
the major health concerns in the United States imposing a huge economic load
on individual/public health care costs, leading to prolonged disabilities
and decreased productivity of our workforce, with further socio-economic
impact. Engineering/Science students will be recruited for this research and
findings incorporated into a course on "Atomistic Simulation of Materials"
at Caltech. |
| |
|
| |
| Reports |
 | Private |
|
| Related Publications |
 | Jaramillo-Botero et al., First-Principles Based
Approaches To Nano-Mechanical And Biomimetic Characterization Of
Polymer-Based Hydrogel Networks For Cartilage Scaffold-Supported
Therapies. Submitted to Special Issue of Journal of Computational and
Theoretical Nanoscience, Computational and Theoretical Nano-Materiomics:
Properties of Biological and de Novo Bioinspired Materials, 02-2009. |
 | Mechanical and transport properties of the
poly(ethylene oxide)-poly(acrylic acid) double network hydrogel from
molecular dynamic Simulations (vol 111B, pg 1729, 2007), Jang et al, J.
Phys. Chem. B 111 (51): 14440-14440 (2007) |
|
| Internal Information |
|
| |
|
| This material is
based upon work supported by the National Science Foundation under Grant No.
0727870. Any opinions, findings, and conclusions or recommendations
expressed in this material are those of the autohor/s and do not necessarily
reflect the views of the National Science Foundation. |
 |
|
|
|
Samsung
Electronics GRO |
2010 |
|
Point of
Contact |
|
|
Start date |
September 1, 2010 |
|
Expires |
TBD |
| |
|
|
Investigators |
|
| Abstract |
The
Humane Genome Project,
the Personal Genome Project,
and the X-prize foundation, among
others, have catalyzed interest in the development of high-speed,
high-throughput, cost-efficient DNA sequencing methods. The race to enable
full-genome sequencing in hours for less than US$1,000, via ultra high
throughput arrays of nano-devices capable of concurrently reading unlabelled
ssDNA at ~1M bases/sec is on. Full-genome sequencing with current
technologies (e.g. chemical- or enzymatic-based shot-gun DNA sequencing,
including massively parallel automated sequencers based on slab gel
separation or capillary electrophoresis) are far from reaching the target
throughput at the target costs. On the other hand, Nanotechnology-based
sequencing devices for Kilobase length (~50,000 or more bases) unamplified,
unlabeled, single-stranded genomic DNA or RNA are, in principle, capable of
the required throughput and cost, yet, no single device has been
demonstrated to this end.
We believe that a DNA nanosequencing device will most likely
involve a nano-fluidics solid-state semiconductor chip consisting of a nano-pored
membrane separating two chambers containing aqueous electrolytes and probes
for electrophoretically controlling the ssDNA translocation across the
membrane and for uniquely distinguishing the physical/electronic signatures
of each nucleotide in a strand sequence. DNA preparation would involve
isolation, purification and concentration in one of the pH and temperature
controlled reservoirs of the nanodevice of ~700mg, high molecular weight
(>50,000 base-pair fragments) human diploid genomic DNA material.
A critical factor to the realization of these devices
involves designing and understanding the precise mechanisms for controlling
the ionic currents and intra-pore fields to serially translocate the ssDNA/RNA
through the electrophoretic nanopores in a membrane with electronically
addressable electrodes, and the nucleotide sequence reading with single base
accuracy at the required throughputs. We propose to establish the most
viable mechanisms for candidate DNA translocation and nucleotide-reading
nano-devices based on solid-state voltage-biased nanopores (or nanochannels)
in membrane-separated electrolyte environments, using a
first-principles-based multiscale modeling approach. |
 |
| |
| Objectives |
Confidential |
| |
| Reports |
 | Confidential |
|
| Related Publications |
|
| Internal Information |
|
| |
|
| |
|


![]()